An overview of the design and conduct of the BATTLE trials
Introduction
Lung cancer is the leading cause of cancer-related mortality in the United States and the world. It accounts for more deaths each year than the combined deaths resulting from breast, colon, prostate, liver, and kidney cancers (1). Non-small cell lung cancer (NSCLC), one of the two major forms of lung cancer, accounts for about 85% of all lung cancers. It is often diagnosed at an advanced stage. Systemic chemotherapy is currently the mainstay for treating metastatic lung cancer. In recent years, targeted agents have been developed for selected patient populations that are more effective and less toxic than conventional chemotherapy. Examples of such agents are erlotinib and gefitinib, which are tyrosine kinase inhibitors (TKIs) of the epidermal growth factor receptor (EGFR). These agents are used in patients who have NSCLC and mutated EGFR (2-5).
Recent advances in biomedicine and genomics have brought better understanding of cancer-causing mechanisms and the ability to identify the corresponding therapeutic targets. Pharmaceutical companies and research institutions are working diligently to screen a myriad of compounds and their combinations that have the potential to address these therapeutic targets and achieve clinical benefits (6). The time and resources devoted to drug development are enormous. However, specific targeted agents may not benefit the general population of patients but work for only a small proportion of patients, and some agents may not work well at all. Therefore, modern drug development involves not only testing the targeted agents for their treatment benefit, but also requires the identification of the target patient population with the corresponding predictive markers.
Challenges exist in the discovery, testing, validation, and functional investigation of the co-development of targeted therapies and their corresponding predictive markers. First, the predictive markers that correspond to the targeted therapies are often unknown at the beginning of a trial. Hence, methods need to be developed to select markers by carefully sieving through a large number of candidate biomarkers for discovery and validation. Second, finding the optimal strategy for testing the treatment effect is not a trivial matter: investigators must determine whether the targeted treatment should be tested first in the unselected population or in the selected population. That task is even more complicated when there are multiple agents with multiple putative markers to be developed. It can be difficult to efficiently pair the agents and biomarkers in a clinical trial when the properties of neither are well understood. Third, in order to match biomarkers and treatments, the biomarker assay has to be done in real time in a reproducible environment. Furthermore, as biomarker analyses are often based on the original tissues removed at the time of diagnosis because that is the only tissue available, they may not accurately reflect the current status of the disease. For example, when patients experience cancer recurrence, they have likely received several lines of therapy; therefore, any biomarker findings for such patients that are based on tissues removed prior to those treatments may or may not reflect the biomarker status of their recurrent tumor.
In view of these challenges, it is desirable to use a trial design that is adaptive so the conduct of the trial can be modified on the basis of cumulative information learned from the trial. For example, adaptive randomization allows for a higher probability that more patients will be assigned to better treatments based on the cumulative outcome and biomarker data. Assigning more patients to more effective treatments based on the corresponding predictive markers not only enhances the individual ethics of the trial, but also improves the accuracy in estimating the treatment effects in such a setting because of the increased sample size in the matched groups. Interim monitoring of the trial with an early stopping rule can stop patient enrollment for clear findings of efficacy, lack of efficacy, and/or unacceptable toxicity. Seamless phase I/II or phase II/III trials can shorten the development time by removing the “white space” between trial phases. Adaptive trial designs are promising in identifying useful predictive markers and effective therapeutic agents in an efficient way while providing the best available treatments to patients during the study (7-10).
The novel phase II Biomarker-integrated Approaches of Targeted Therapy for Lung Cancer Elimination (BATTLE) program described in this article consists of the BATTLE-1 trial—the first completed, prospective biopsy-mandated, biomarker-based, adaptively randomized phase II clinical trial in patients with previously treated NSCLC (11,12)—as well as the subsequent BATTLE-2 trial (13). We have demonstrated the feasibility of what was previously thought to be an impossible task: acquiring tumor tissues in patients with recurrent lung cancer and subjecting them to real-time biomarker analysis (14). The success of this program has opened a new era of targeted agent testing that is integrated with discovering and validating novel markers and offering better treatments for patients enrolled in the trials. This program sets an excellent example for the design and conduct of clinical trials that implement Bayesian adaptive designs in the development of targeted therapy. It is a step toward achieving personalized medicine.
The rest of this paper is organized as follows. In sections 2 to 5, we describe the design, conduct, and results of the BATTLE-1 trial, as well as the lessons we learned from that process. In section 6, we present additional publications and work related to the BATTLE-1 trial. In section 7, we describe the BATTLE-2 trial. In section 8, we summarize the impact of the BATTLE trials and conclude with a brief discussion on the future direction of related research.
Design of the BATTLE-1 trial
The concept of the BATTLE-1 trial was initially discussed in 2005. The BATTLE-1 program consisted of one umbrella trial and four parallel phase II studies with biomarker-based, targeted therapies in patients with advanced NSCLC who had been previously treated with chemotherapy and subsequently experienced disease relapse. Figure 1 shows the BATTLE-1 schema. The four treatment arms were erlotinib, sorafenib, vandetanib, and the combination of erlotinib and bexarotene. The treatments were chosen to target each of the four selected gene pathways in NSCLC that were of the highest scientific and clinical interest at the time when the trial was designed. It was assumed that each treatment could be more efficacious in patients with a certain biomarker profile that matched the agent’s mechanism of action.
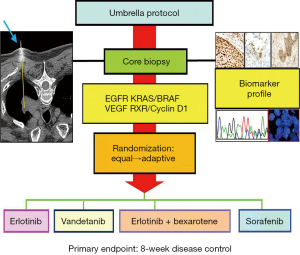
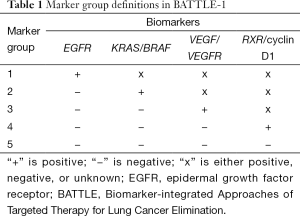
BATTLE-1 was a biopsy-mandated study. Eligible patients gave their consent to undergo a tissue biopsy before they were treated. A core-needle biopsy, guided by computed tomography or ultrasound, was used to collect tissues for the required biomarker analysis and the additional gene expression, mutation, and proteomic biomarker analysis. A patient’s treatment assignment was based on his or her biomarker profile, which was defined by eleven pre-specified biomarkers: EGFR mutation, EGFR overexpression/amplification, EGFR increased copy number (in the EGFR pathway), KRAS mutation, BRAF mutation (in the KRAS/BRAF pathway), VEGFR expression, VEGFR-2 expression (in the VEGFR pathway), RXRα expression, RXRβ expression, RXRγ expression, and cyclin D1 expression (in the RXR/cyclin D1 pathway). The screening of these eleven individual markers meant that we could have had 2,048 possible marker combinations, even from simply dichotomizing each marker as positive or negative. To reduce the number of parameters, we sequentially examined the presence or absence of certain biomarkers to classify each patient into one of the five marker groups listed in Table 1. For example, if any of the biomarkers related to the EGFR pathway were positive for a given patient, the patient was classified into the EGFR marker group (markergroup 1) regardless of the status of the other biomarkers for that patient. Otherwise, if the patient’s tumor sample showed KRAS or BRAF mutations, the patient was classified into the KRAS/BRAF marker group (marker group 2) regardless of the status of the remaining biomarkers for that patient, and so forth. If none of the pre-specified biomarkers were positive for a given patient, the patient was classified into the fifth marker group, which included patients for whom the biomarker information was missing or incomplete.
Full table
The goal of the BATTLE-1 trial was to establish a clinical trial platform that advanced trial design in the development of targeted therapies, and to use the biomarker data to assess the clinical benefit of targeted molecular agents in patients with advanced NSCLC. Specifically, we aimed to provide an accurate estimate of the true disease control rate (DCR) for each of the treatment arms in each of the marker groups. In addition, the trial design was adaptive so that it assigned more patients to the more promising treatment arms based on data accumulated in the trial up until that time according to each patient’s biomarker profile. Conversely, the trial suspended patient enrollment in the ineffective treatment arms early based on the patient’s biomarker profile.
The 8-week DCR was chosen as the primary endpoint to use in evaluating the treatment effect. It was an easily and quickly assessable endpoint that had been shown to be a reasonable surrogate for the overall survival time in patients with advanced lung cancer (15). In order to simultaneously evaluate the four treatments and five marker groups and to identify the most efficacious treatment in each marker group, a Bayesian hierarchical probit model was applied (11). This model allowed for borrowing statistical strength among the five marker groups within the same treatment arm, which can improve the accuracy of estimation if patients from different marker groups who receive the same treatment show similar treatment responses.
An outcome-adaptive randomization scheme was employed in the BATTLE-1 trial. Eligible patients were first equally randomized into each of the four treatment arms based on their marker group membership. Once at least one patient had been treated in each of the 20 treatment-by-marker subgroups, adaptive randomization began. Patients were adaptively randomized into the treatment arms in proportion to the estimated posterior DCRs within each biomarker group. Adaptive randomization allowed us to learn the performance of each treatment arm in each marker group during the trial, and to use the updated knowledge to guide the assignment of patients to treatment arms as the trial continued. As a result, more patients received the more efficacious treatments as the study progressed.
In the BATTLE-1 trial, we applied the Bayesian adaptive design to continuously update and learn from the information and to perform interim monitoring for futility. The Bayesian framework allows for the natural implementation of an early stopping rule such that the assignment of patients in a particular marker group to a given treatment arm can be suspended if the treatment is found not to be promising for that marker group. A not-promising treatment was defined as one that had a likelihood of its estimated DCR being higher than 50% (and the targeted DCR or the DCR under the alternative hypothesis) was lower than 10%. At the end of the study, we declared a treatment as successful in a given marker group if the probability of the estimated DCR exceeding the historical threshold of 30% (the DCR under the null hypothesis) is greater than 80%. Before we applied the design in the BATTLE-1 trial, we conducted extensive simulation studies to evaluate the performance of the design under various scenarios. The probability cutoffs were calibrated so that the type I and type II error rates were well controlled. When a treatment-by-marker subgroup had a true DCR of 30%, the probability of it being declared a success was 20% or less (type I error). If a treatment-by-marker subgroup had a true DCR of 60%, the probability of declaring a treatment a success was at least 80% (statistical power). The probability of declaring a treatment a success can be as high as 95% when the DCR is 80%. A high type I error rate was selected in order to increase the statistical power such that we would have a high probability of selecting a potentially efficacious treatment and a low probability of overlooking a potentially efficacious treatment.
In contrast to the traditional single-arm design for phase II studies, which would have involved 20 separate parallel studies to evaluate the efficacy of four treatments in five biomarker groups, the Bayesian adaptive design allowed us to enroll patients under one study. The use of a hierarchical design and early stopping rules for futility improved the efficiency of the study. The outcome-adaptive randomization scheme enhanced the individual ethics of the trial and patient comparability across the different treatments. The adaptive design which puts all patients under one roof also enhances the patient comparability in contrast to the sequentially conducted multiple single-arm phase II trials.
Conduct of the BATTLE-1 trial
With four treatments, five marker groups, real-time biomarker analysis, and a Bayesian adaptive design, it was a logistically challenging task to conduct the BATTLE-1 trial efficiently and effectively. To facilitate the conduct of the trial, we built an integrated web-interfaced database application. Figure 2 illustrates the trial conduct and the associated web application. All of the information about the patients and the study was stored in an electronic database. The information for each patient was carefully recorded from the day the patient registered for the study to the day the patient completed the study. An eligible patient was registered and then evaluated with a baseline physical exam, lab test, mandated biopsy, molecular pathology assessment, and biomarker analysis to determine the appropriate marker group. By design, the goal was to perform molecular testing within 2 weeks of the tissue biopsy. Then, adaptive randomization was performed by calling an R computer code through the web services to generate an assignment for the patient to a treatment arm. Regular clinical visits took place during the initial 8-week treatment period and patient compliance and any adverse events related to the treatment were recorded. The primary endpoint of disease control was evaluated at 8 weeks after randomization. The radiographic measurement of tumor size and the clinical outcomes were recorded to determine the tumor response. Information obtained at subsequent follow-up visits was also recorded until the patient went off study due to either disease progression, experiencing toxicity, or loss to follow-up. All data were stored within CORe, the MD Anderson regulatory environment, and the study-specific SQL 2005 database. Reports were generated periodically to monitor the study progress.
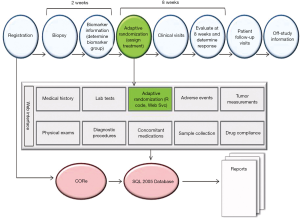
A requirement of the Bayesian adaptive trial design is timely measuring and reporting of the study outcomes such that the randomization probability and the posterior probability for futility monitoring can be calculated accurately on the basis of the most recent data. Whenever a patient’s disease control status was updated, the posterior distribution of the estimated DCR was calculated and updated accordingly. The updated information was used to compute the randomization probability and check whether the early stopping boundary for futility had been reached for certain treatments in certain marker groups. If the early stopping boundary for futility were reached, patient randomization would be suspended for that treatment in that marker group. All these computationally intensive calculations were performed in R code automatically and assessed through the web services. This adaptive learning and dynamic treatment allocation very nicely illustrated the motto of Bayesian adaptive design: “We learn as we go”. To meet the timeliness requirement of measuring and entering the 8-week disease control status, an automatic e-mail notification system was developed. It was programmed to send an e-mail to the designated research coordinator to remind the coordinator to schedule a patient visit when6 weeks had passed since the patient had been randomized. The system also kept track of the time when the 8-week endpoint was recorded and automatically sent e-mail alerts when an endpoint evaluation was overdue for more than2 weeks.
To accurately evaluate the treatment outcome, an endpoint review committee was formed that included clinicians, radiologists, and research nurses. The committee reviewed the treatment outcomes during and at the end of the study to ensure consistent criteria were followed while blinded to the patient’s treatment assignment. During the trial, automatic alerts were sent to the appropriate personnel to alert them to a delayed response entry, suspension of patient accrual to certain treatments for a subgroup, or other unexpected or adverse events.
The conduct of the BATTLE-1 trial required substantial teamwork and collaboration. It involved the creation of an integrated multidisciplinary research team of clinicians who evaluated and treated the patients, interventional radiologists who performed the image-guided core-needle biopsy, pathologists and basic scientists who performed the histology reading and biomarker analyses, statisticians who provided the trial design and implemented the algorithm for adaptive randomization, pharmacists who dispensed study medicines, radiologists who evaluated the tumor response, research nurses and research coordinators who worked with patients step-by-step during the entire trial period, and computer programmers who built and maintained the web-interfaced database applications. Everyone in the team worked together to ensure the smooth conduct of the study. Although much effort was required to build and operate a multidisciplinary team for conducting an adaptive trial, as a result, the BATTLE-1 trial was implemented exquisitely and high quality data were collected throughout the trial.
Results of the BATTLE-1 trial
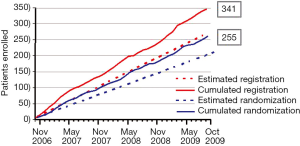
The BATTLE-1 trial was activated in November 2006, and patient accrual was completed in October 2009. In those 3 years, a total of 341 patients were enrolled in the study. Among that total enrollment, 255 patients were randomized to the treatment arms and 86 patients were not randomized because of either concurrent illness, worsening overall condition, a condition preventing a biopsy, or the choice of the patient or the treating physician to seek alternative treatments. Figure 3 shows the accumulating number of patients enrolled and randomized in the trial over time. The patient accrual rate was about 9.5 patients per month, which was better than the expectation of eight patients per month. On average, 7.1 patients were randomized each month. Both clinicians and patients were enthusiastic to participate in the study. The concepts of personalized medicine and adaptive trial designs were well accepted by the clinicians and patients.
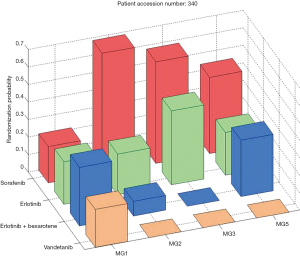
Among the patients who were randomized to treatments within the trial, 244 had an evaluable 8-week disease control status. The overall 8-week DCR was 46%. The marginal DCRs were 34%, 33%, 50%, and 58% for the treatments of erlotinib, vandetanib, erlotinib plus bexarotene, and sorafenib, respectively. The adaptive randomization scheme assigned the most patients (n=105) to receive sorafenib because it had a better marginal DCR compared to the other three treatments (erlotinib: n=58; vandetanib: n=52; erlotinib plus bexarotene: n=36). Figure 4 shows the distributions of the final randomization probability into the four treatments for marker groups 1, 2, 3, and 5 (marker group 4 is not shown because only six patients belonged to it). Confirming the initial hypothesis, the trial showed that patients in the KRAS/BRAF marker group had a much higher DCR (79%) when treated with sorafenib, compared to the DCRs of 14% for erlotinib, 0% for vandetanib, and 33% for the combination of erlotinib and bexarotene. In addition, erlotinib plus bexarotene worked well in the RXR/cyclin D1 marker group. We also performed exploratory analyses to identify potential predictive biomarkers. The DCR for patients in the KRAS mutation group was higher when treated with sorafenib compared to erlotinib (61% vs. 22%). Though erlotinib did not show significantly high DCRs among patients in the EGFR marker group, it did have a higher DCR for patients with the single marker of EGFR mutation compared to those with the wild-type EGFR (71% vs. 29%). Of interest, patients with wild-type EGFR had a better DCR when treated with sorafenib than with erlotinib (64% vs. 29%). More complete results can be found in the original publication of the BATTLE-1 trial (12).
Lessons learned from the BATTLE-1 trial
The BATTLE-1 trial was the first completed, prospective biopsy-mandated, biomarker-based, adaptively randomized clinical trial for patients with relapsed NSCLC. Compared to using tissue samples and biomarker status assessed at the time of diagnosis, the re-biopsy in patients with disease relapse and the real-time biomarker analysis provided an accurate biomarker status for the current treatment assignment in the trial and a wealth of information for future studies. By using a fresh core-needle biopsy, not only did we obtain the tissue samples needed to define the patient’s biomarker profile for treatment assignment in the BATTLE-1 trial, but we procured tissue samples that will be available for future studies. In addition, from patients who consented, we collected blood and serum samples at baseline and after treatment. All this information will enable us to discover and validate novel biomarkers in future studies.
The BATTLE-1 study confirmed our pre-specified hypotheses that patients with EGFR mutations had better disease control when treated with erlotinib and patients with KRAS/BRAF mutations had better disease control when treated with sorafenib. The study also identified some interesting findings, for example, the predictive effects of better DCR for patients with KRAS mutations or wild-type EGFR who were treated with sorafenib, for patients with high VEGFR-2 expression who were treated with vandetanib, and for patients with high cyclin D1 expression who were treated with erlotinib plus bexarotene. Of course, all these findings are based on small sample sizes and therefore must be validated in future BATTLE trials and in other studies.
The BATTLE-1 trial used a Bayesian adaptive design. Compared to traditional equal randomization or fixed rate randomization schemes, the outcome-adaptive randomization scheme allows us to adjust the randomization probability as the data accumulate during the trial. By using the accumulating data, our knowledge of the treatment effect can be continuously updated during the trial. Consequently, future patients can be assigned to better treatments with higher probability according to their biomarker profiles. Thus, the design enhances individual ethics. This adaptive feature not only refines our initial assumption of the treatment effect, but, should our initial assumption be wrong, this feature can correct the assumption as the data accumulate such that the amount of information from the trial overwhelms the prior information. Better estimation of the treatment effect can be achieved when a larger sample size is achieved by assigning more patients to more effective treatments in patients with the corresponding predictive markers. As shown, some findings from the BATTLE-1 trial have validated ourpre-specified scientific hypotheses regarding biomarkers that are predictive of disease response to targeted agents and, more importantly, the trial has also identified potential new predictive markers to be studied in the future.
The successfully conducted BATTLE-1 trial has made important clinical discoveries and demonstrated the feasibility of its novel design for advancing personalized treatment in NSCLC. It also leaves room for improvement in the future. Notably, in the BATTLE-1 trial, the biomarkers were pre-specified in the study based on our experience and the research literature available at that time. However, some of the selected biomarkers did not have any observable prognostic or predictive effects, e.g., RXRs. Furthermore, although the biomarker grouping reduced the number of parameters in the model and simplified the trial design, combining markers weakened the association between the real predictive biomarkers and the treatments. For example, we formed the EGFR marker group from three subgroups: EGFR mutation, EGFR overexpression/amplification, and EGFR increased copy number. The predictive effect of the EGFR mutation with the erlotinib treatment was very strong but was diluted after grouping it with EGFR overexpression/amplification and EGFR increased copy number. We have learned that it is not a good idea to pre-select the study markers, particularly in the setting when little is known about the new treatments and their corresponding markers. We also learned that grouping different genetic mutations or characteristics to form fewer marker groups is not desirable because the true marker effect can be weakened by incorporating unimportant markers.
Equal randomization was applied in the first stage of the trial to gather the information required to form the prior distribution that would be used for adaptive randomization in the second stage. We stipulated that the adaptive randomization scheme would start after we enrolled at least one patient in each of the marker-by-treatment subgroups. It turned out that few patients belonged to the RXR/cyclin D1 marker group, which unfortunately delayed the start of the adaptive randomization scheme until about 40% of the patients had been equally randomized to the various treatments. Looking back, we determined that we should have allowed the adaptive randomization scheme to start earlier, say, after about 20% to 25% of the patients had been equally randomized, so that more patients could have benefited from adaptive randomization.
Another hurdle that inadvertently impacted the adaptive randomization scheme in the BATTLE-1 trial involved the eligibility criteria specific to each treatment. The unique properties of each treatment required the use of treatment-specific eligibility criteria in addition to the eligibility criteria common to all trial participants. Patients enrolled in the BATTLE-1 trial had advanced stage NSCLC and therefore had already received cancer treatments, typically two to six lines of treatment. Their resulting medical conditions disqualified many of the patients from eligibility for all of the BATTLE-1 treatments. In fact, only 14% of the patients were eligible for all four treatments in the trial. Patients can only be randomized among the treatments for which they are eligible; thus, for 86% of the patients enrolled in the trial, we had to adjust the adaptive randomization according to the patient’s eligibility.
It is well known that adaptive trial designs are prone to experience a study population drift (16). The study population in the BATTLE-1 trial was quite stable in general; however, over the course of the study, we found that more smokers and patients who had previously received erlotinib enrolled in the latter part of the study compared to the beginning of the study. Statistical methods such as covariate adjusted regression analysis are available to alleviate the impact of an unbalanced covariate distribution during a trial. Adaptive randomization works best when used with effective treatments and markers that show good predictive performance. The final lessons learned were that some of the pre-specified markers were not predictive of the treatment response and some of the treatments were not as successful as we anticipated, and these factors limited the success that could be achieved in the trial.
Additional publications from the BATTLE-1 trial
The BATTLE program and the first completed trial compose a rich learning environment through which we have explored many topics in NSCLC research, medical practice, clinical trial design and conduct, and the continuing development of novel statistical methods in medical research. Here, we report selected publications from the BATTLE-1 trial. Ihle et al. conducted microarray analysis of mRNA expression on frozen core biopsy tumor samples from the patients who participated in the BATTLE-1 trial (17). They found that patients who had either mutant KRAS-Gly12Cys or mutant KRAS-Gly12Val had worse progression-free survival compared with patients who had other mutant KRAS proteins or wild-type KRAS. Tsao et al. performed an analysis that focused on elderly patients (18). Of interest, they found that elderly men showed better clinical benefit from certain targeted agents. For example, men aged 65 to 70 years had better progression-free survival when treated with vandetanib, and men over 70 years of age had better progression-free survival when treated with sorafenib. Tam et al. assessed the acquisition of tissue for biomarker analysis using the image-guided percutaneous transthoracic core-needle biopsy and determined that the success rate for obtaining tissue was 82.9% in patients in the BATTLE-1 trial (14). Byers et al. developed and validated a 76-gene epithelial-mesenchymal transition (EMT) signature using gene expression profiles from four microarray platforms of NSCLC cell lines and patients treated in the BATTLE-1 trial (19). The EMT signature predicted resistance to EGFR and PI3K inhibitors and identified Axl as a potential therapeutic target for overcoming resistance to EGFR inhibitors.
Gene expression and the biomarker effect in each of the four targeted agents in the BATTLE-1 trial have been further studied. For example, Tsao et al. reported that vandetanib improved progression-free survival in patients with EGFR mutation compared to patients with wild-type EGFR, if the patients’ tumors were resistant to EGFRTKIs (20). For patients treated with sorafenib, three important findings were documented: (I) significant clinical benefit for those with mutated KRAS versus wild-type KRAS; (II) significant clinical benefit for those with wild-type EGFR versus mutated EGFR; and (III) the gene expression profiles from NSCLC cell lines and patient tumor biopsies with wild-type EGFR were used to develop a sorafenib sensitivity signature that showed improved progression-free survival among patients with wild-type EGFR (21).
Related research conducted outside of the BATTLE team by Dragnev et al. showed that bexarotene plus erlotinib suppressed lung carcinogenesis independent of KRAS mutations in clinical trials and transgenic mouse models (22). Cotargeting cyclin D1 via the retinoid X receptor and EGFR by combining erlotinib and bexarotene is a potentially promising venue for the prevention and treatment of lung cancer (23). In addition, knowledge gained from the mechanistic approach to treating lung cancer in the BATTLE program led to the proposal of the concept of reverse migration as a new strategy for personalized lung cancer prevention (24).
From the statistical methodology point of view, the BATTLE program has inspired the development of Bayesian adaptive trial designs and the evaluation of various trial designs for studying targeted agents (25). Outcome-adaptive randomization has been shown to be very useful when a large difference in efficacy is found among treatments or when the goal is to maximize the overall treatment benefit for patients enrolled in the trial, particularly when the applicable patient population beyond the trial is small (26). Furthermore, in order to select relevant prognostic and predictive markers, a Bayesian 2-step Lasso strategy with a group Lasso approach followed by an adaptive Lasso approach was developed for time-to-event endpoints (27).
Extension of the BATTLE-1 trial: the BATTLE-2 trial
The BATTLE-1 trial demonstrated a new platform for novel adaptive clinical trial design and has allowed the investigators to derive interesting findings for validation in future studies. Major limitations of the BATTLE-1 trial were the pre-selection of biomarkers and bundling the biomarkers into marker groups. To rectify this problem, we have designed a Bayesian 2-stage biomarker-based adaptive randomization trial called BATTLE-2 (13). The BATTLE-2 trial has been designed for the same patient population that was eligible for the BATTLE-1 trial, and uses the same primary endpoint, i.e., the 8-week DCR, which has been shown to be a good surrogate of the overall survival time in BATTLE-1 and other studies (7,8). In BATTLE-2, four treatments were selected: erlotinib (serving as the control group), sorafenib, MK-2206 (an AKT inhibitor) plus erlotinib, and MK-2206 plus AZD6244 (a MET inhibitor). The study schema is shown in Figure 5.
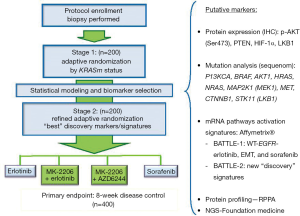
The potential prognostic/predictive biomarkers are identified during the training phase (pre-BATTLE-2) on the basis of prior studies and the literature. These putative markers are tested in the first stage and validated in the second stage of the BATTLE-2 trial. The trial is designed to achieve three goals: (I) test the treatment efficacy of the targeted agents and their combinations; (II) identify the corresponding prognostic and predictive markers; and (III) treat patients with the most effective treatment in the study based on the available data. Adaptive randomization is applied in both stages to assign more patients to better treatments based on the individual patient’s biomarker profile. In contrast to the BATTLE-1 trial, adaptive randomization in BATTLE-2 starts at the beginning of the study stratified by the KRAS mutation status. Note that the randomization probability is set to be bounded between 0.2 to 0.8 to ensure that the AR allows patients to be assigned to all treatment arms with reasonable probabilities in stage 1. A “Go or No-Go” decision is made at the end of the first stage by testing the treatment effect of each individual treatment. If none of the experimental treatments shows any promising effect compared to erlotinib (the control group) in all patients and in any marker subgroup (wild-type or mutated KRAS), a “No-Go” decision is rendered and the trial can be stopped early. On the other hand, if a “Go” decision is made at the end of the first stage, the process of biomarker analysis and selection is implemented to screen for and select additional prognostic or predictive biomarkers based on the data from the first stage and other available information. A refined predictive model is used for the adaptive randomization scheme in the second stage. Note that the randomization probability is set to be bounded between 0.1 to 0.9 to guard against extreme allocation in stage 2.
The plan is to enroll a total of 400 evaluable patients over a 4-year period. With a conservative estimate that 10% of the patients may have incomplete marker profiles due to limited numbers of tumor cells in the biopsy samples or an unevaluable endpoint, a total of 450 patients will be enrolled. Simulations were applied to thoroughly study the operating characteristics of the design, with the goal of achieving at least 80% power at a 10% type I error rate for testing the efficacy of each treatment, as well as yielding at least 80% power for identifying important prognostic and predictive markers.
Patients with a prior history of having received erlotinib treatment are not randomized into the erlotinib-only treatment arm. Treatment effects are tested separately in the patient subgroups stratified by whether or not patients had prior erlotinib exposure as well as in the overall patient groups. As mentioned previously, BATTLE-2 uses a 2-stage design. In the first stage, patients are adaptively randomized based on their KRAS mutation status and whether they were previously treated with erlotinib. An early futility stopping rule is activated from the 71st patient. If all the experimental treatment arms do not show evidence of improved efficacy over the control arm (erlotinib) for all patients and any of the biomarker groups (KRAS mutation negative or positive) by prior erlotinib treatment status, the trial is stopped early. By the end of stage 1, if the trial has not been stopped, then the biomarker analysis is performed through a training, testing, and validation procedure described as follows.
Before BATTLE-2, over 100 discovery biomarkers were screened to identify putative prognostic and predictive markers. Combining with the finding in the BATTLE-1 trial, promising prognostic and predictive markers are identified in the training step. During stage 1 of the trial, the identified markers are assessed in the patients’ tissues and blood samples. Those data, as well as the patients’ medical demographic variables and treatment outcomes, supplemented by other up-to-date in vitro or in vivo data and information from the literature, are combined by the biostatistics and bioinformatics team to propose a refined predictive model to be tested. We test the “best-performing” markers from the data in the first stage of BATTLE-2. For markers passing the training and testing steps, they will be further validated in the second stage of BATTLE-2. The predictive markers identified in stage 1 are used for adaptive randomization in stage 2. We apply the Bayesian 2-step Lasso method for variable selection at the end of stage 1. Specifically, the first step of variable selection is a group selection procedure aimed at identifying markers with either prognostic effects or predictive effects. The second step is an individual selection for a marker and its interactions with the treatments. The final decision of biomarker selection is based on statistical strength, biological plausibility, and practical considerations. Upon final selection of markers and the refined predictive model, we amend the protocol for IRB approval and continuously adaptively randomize patients in stage 2 if the trial is not stopped early. By the end of the study, all markers will be evaluated for potential predictive or prognostic effect, and the effective treatment in patients with predictive markers will be declared.
Using the experiences and knowledge gained from the conduct of the BATTLE-1 trial, the BATTLE-2 trial has been designed with more flexibility: no restriction of pre-specified biomarkers, no biomarker grouping, adaptive randomization starting from the beginning of the trial, prognostic/predictive biomarkers being screened and selected in a 3-step process: training, testing, and validation, and the predictive model for adaptive randomization being refined with real-time data observed for the study. Due to its exploratory nature, BATTLE-2 has received the investigational device exemption (IDE) waiver after a meeting with the FDA in January 2013. Like BATTLE-1, BATTLE-2 is being conducted through a web-based application. The first stage of patient accrual was opened at the UT MD Anderson Cancer Center and the Yale Cancer Center in June 2011. Patient accrual and biomarker analyses have continued in the subsequent years.
Impact of BATTLE trials and future directions
The BATTLE program has demonstrated the feasibility and impact of the first biomarker-based, adaptively randomized novel clinical trial platform in NSCLC, and has set an example for the development of targeted agents in cancer. The successful conduct of this program has demonstrated that it is feasible in modern medical practice to undertake real-time biomarker analysis following a tissue biopsy in patients with relapsed disease. The primary paper describing the BATTLE-1 trial (12) has been cited in more than 200 articles and book chapters to date. The successful completion of that study has been called “an important milestone” in the war against cancer (28). In NSCLC diagnosis and treatment, BATTLE-1 is a landmark trial for successfully pairing biomarker-defined cohorts of patients with targeted therapeutics (29). The completion of BATTLE-1 has proven that we can expand the horizon of oncology clinical trial research to incorporate a prospective biopsy and real-time biomarker analysis. This alleviates many problems such as selection bias and the inflation of the type I error rate in retrospective studies based on post-treatment subgroup analysis. It also addresses the problems of biomarker assays obtained from the original diagnostic tissue, which is far from satisfactory because of the changes that may occur in a patient’s biomarker profile after the patient receives many lines of treatment. It can help us to achieve a more accurate understanding of the cancer-causing mechanism, to efficiently identify the predictive biomarkers and corresponding targeted therapies, and to use this information to provide better treatment for patients enrolled in the trial.
Applying Bayesian adaptive designs in the BATTLE trials provides excellent examples of how to fill the gap between statistical methodology research and its application in medical practice. Though more researchers are realizing the advantage of using Bayesian adaptive clinical trial designs, real applications of such designs in clinical trials are still limited. It is a common occurrence for there to be a long time lag between the publication of a statistical method and its application in a clinical study. For example, the seminal continuous reassessment method for phase I trials was published in 1990 (30), but was not widely used for some time. Reviewing the Science Citation Index database between 1991 and 2006, it was found that only 1.6% of the 1,235 phase I trials reported used Bayesian adaptive designs (31). A recent review of published Bayesian adaptive clinical trials indicated that the challenges when using Bayesian adaptive trial designs were the difficulties of the Bayesian computations and the lack of user-friendly software for the study design and trial conduct (32). However, more and more tools have been developed in recent years to conquer these computational barriers when using Bayesian methods (33). One notable example is the collection of useful software that is available at the UT MD Anderson Cancer Center software download site (https://biostatistics.mdanderson.org/SoftwareDownload/). The successful conduct of the BATTLE trials and studies that have similarly applied Bayesian adaptive designs, such as the I-SPY2 trial (34), has promoted methodological research in novel clinical trial design and encouraged statisticians and clinical trialists to implement more new design methods in their medical research (35). Concurrently, there have been several major attempts to apply similar concepts in the quest to identify effective cancer therapies and associated predictive markers. These include the National Cancer Institute’s Molecular Analysis for Therapy Choice Program (MATCH; http://www.cancer.gov/clinicaltrials/noteworthy-trials/match#match), the Lung Cancer Master Protocol (Lung-MAP; http://www.cancer.gov/newscenter/newsfromnci/2014/LungMAPlaunch), and the Adjuvant Lung Cancer Enrichment Marker Identification and Sequencing Trials (ALCHEMIST; http://www.cancer.gov/clinicaltrials/noteworthy-trials/alchemist).
The BATTLE program has created a new paradigm of prospective biopsy-based, real-time biomarker analysis and adaptive designs in clinical studies. With advancements in biomedical research, we look forward to more such studies increasing trial efficiency and enhancing the benefit to patients while developing more effective treatments. The BATTLE program has opened a new page for clinical trial design and conduct, and has brought us one step closer to personalized medicine.
Acknowledgements
Funding: This work was supported in part by the grant CA016672 from the National Cancer Institute. The BATTLE-1 research was supported in part by the Department of Defense Grant W81XWH-6-1-0303. The BATTLE-2 research was supported in part by grant CA155196 from the National Cancer Institute. The clinical trial was supported in part by Merck Research Laboratories and Bayer HealthCare. The authors thank Ms. Lee Ann Chastain for editorial assistance.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008;359:1367-80. [PubMed]
- Kim ES, Hirsh V, Mok T, et al. Gefitinib versus docetaxel in previously treated non-small-cell lung cancer (INTEREST): a randomised phase III trial. Lancet 2008;372:1809-18. [PubMed]
- Douillard JY, Shepherd FA, Hirsh V, et al. Molecular predictors of outcome with gefitinib and docetaxel in previously treated non-small-cell lung cancer: data from the randomized phase III INTEREST trial. J Clin Oncol 2010;28:744-52. [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [PubMed]
- Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958-67. [PubMed]
- Tepper RI, Roubenoff R. The role of genomics and genetics in drug discovery and development. In: Willard HF, Ginsburg GS. eds. Genomic and Personalized Medicine. San Diego: Academic Press, 2009:335-56.
- Berry DA. Bayesian clinical trials. Nat Rev Drug Discov 2006;5:27-36. [PubMed]
- Berry SM, Carlin BP, Lee JJ, et al, editors. Bayesian Adaptive Methods for Clinical Trials. Boca Raton, FL: CRC Press, 2010.
- Inoue LY, Thall PF, Berry DA. Seamlessly expanding a randomized phase II trial to phase III. Biometrics 2002;58:823-31. [PubMed]
- Berry DA. Adaptive clinical trials in oncology. Nat Rev Clin Oncol 2011;9:199-207. [PubMed]
- Zhou X, Liu S, Kim ES, et al. Bayesian adaptive design for targeted therapy development in lung cancer--a step toward personalized medicine. Clin Trials 2008;5:181-93. [PubMed]
- Kim ES, Herbst RS, Wistuba II, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov 2011;1:44-53. [PubMed]
- Gu X, Chen N, Wei C, et al. Bayesian Two-Stage Biomarker-Based Adaptive Design for Targeted Therapy Development. Statistics in Biosciences 2014. Available online: http://link.springer.com/article/10.1007/s12561-014-9124-2
- Tam AL, Kim ES, Lee JJ, et al. Feasibility of image-guided transthoracic core-needle biopsy in the BATTLE lung trial. J Thorac Oncol 2013;8:436-42. [PubMed]
- Lara PN Jr, Redman MW, Kelly K, et al. Disease control rate at 8 weeks predicts clinical benefit in advanced non-small-cell lung cancer: results from Southwest Oncology Group randomized trials. J Clin Oncol 2008;26:463-7. [PubMed]
- Karrison TG, Huo D, Chappell R. A group sequential, response-adaptive design for randomized clinical trials. Control Clin Trials 2003;24:506-22. [PubMed]
- Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst 2012;104:228-39. [PubMed]
- Tsao AS, Liu S, Lee JJ, et al. Clinical outcomes and biomarker profiles of elderly pretreated NSCLC patients from the BATTLE trial. J Thorac Oncol 2012;7:1645-52. [PubMed]
- Byers LA, Diao L, Wang J, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res 2013;19:279-90. [PubMed]
- Tsao AS, Liu S, Lee JJ, et al. Clinical and biomarker outcomes of the phase II vandetanib study from the BATTLE trial. J Thorac Oncol 2013;8:658-61. [PubMed]
- Blumenschein GR Jr, Saintigny P, Liu S, et al. Comprehensive biomarker analysis and final efficacy results of sorafenib in the BATTLE trial. Clin Cancer Res 2013;19:6967-75. [PubMed]
- Dragnev KH, Ma T, Cyrus J, et al. Bexarotene plus erlotinib suppress lung carcinogenesis independent of KRAS mutations in two clinical trials and transgenic models. Cancer Prev Res (Phila) 2011;4:818-28. [PubMed]
- Kim ES, Lee JJ, Wistuba II. Cotargeting cyclin D1 starts a new chapter in lung cancer prevention and therapy. Cancer Prev Res (Phila) 2011;4:779-82. [PubMed]
- Gold KA, Kim ES, Lee JJ, et al. The BATTLE to personalize lung cancer prevention through reverse migration. Cancer Prev Res (Phila) 2011;4:962-72. [PubMed]
- Lee JJ. Xuemin Gu, Suyu Liu. Bayesian adaptive randomization designs for targeted agent development. Clin Trials 2010;7:584-96. [PubMed]
- Lee JJ, Chen N, Yin G. Worth adapting? Revisiting the usefulness of outcome-adaptive randomization. Clin Cancer Res 2012;18:4498-507. [PubMed]
- Gu X, Yin G, Lee JJ. Bayesian two-step Lasso strategy for biomarker selection in personalized medicine development for time-to-event endpoints. Contemp Clin Trials 2013;36:642-50. [PubMed]
- Rubin EH, Anderson KM, Gause CK. The BATTLE trial: a bold step toward improving the efficiency of biomarker-based drug development. Cancer Discov 2011;1:17-20. [PubMed]
- Sequist LV, Muzikansky A, Engelman JA. A new BATTLE in the evolving war on cancer. Cancer Discov 2011;1:14-6. [PubMed]
- O'Quigley J, Pepe M, Fisher L. Continual reassessment method: a practical design for phase 1 clinical trials in cancer. Biometrics 1990;46:33-48. [PubMed]
- Rogatko A, Schoeneck D, Jonas W, et al. Translation of innovative designs into phase I trials. J Clin Oncol 2007;25:4982-6. [PubMed]
- Chevret S. Bayesian adaptive clinical trials: a dream for statisticians only? Stat Med 2012;31:1002-13. [PubMed]
- Lee JJ, Chu CT. Bayesian clinical trials in action. Stat Med 2012;31:2955-72. [PubMed]
- Barker AD, Sigman CC, Kelloff GJ, et al. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin Pharmacol Ther 2009;86:97-100. [PubMed]
- Marchenko O, Fedorov V, Lee JJ, et al. Adaptive Clinical Trials: Overview of Early-Phase Designs and Challenges. Therapeutic Innovation & Regulatory Science 2014;48:20-30.


